
Associate Professor
CIHR New Investigator (2012-2017)
MSFHR Career Investigator (2011-2019)
University of British Columbia, 2001, B.Sc.(Honours)
University of British Columbia, 2006, Ph.D.
Harvard Medical School, 2010, Postdoctoral Fellow
Office: Life Sciences Centre, 5301
Office Phone: 604–827–3976
Fax: 604–822–5227
Lab Phone: 604–827–4274
E-mail: calvin.yip@ubc.ca
Website: https://ubcyiplab.wixsite.com/ubcyiplab
Research Interest
1. Autophagy machinery
How does a cell remove and recycle unwanted materials? Autophagy is an evolutionarily conserved pathway that encapsulates large objects to be degraded in a double-membrane vesicle called the autophagosome and targets this cargo to the lysosome for breakdown. Defects in this pathway have been implicated in neurodegenerative disorders, cancers, and other human diseases. We study how a specialized group of proteins that mediate the different steps of this important degradative pathway using biochemical, structural biology, and cell biology approaches.
2. Chromatin modifying complexes
How does a cell establishes and maintains its gene expression pattern and adapts this to different environmental conditions? Eukaryotic genomic DNA exists in a DNA-protein complex known as chromatin. Post translational modifications to the histone proteins that form the nucleosome, the most basic unit of chromatin, is a key mechanism to regulate chromatin structure and gene expression. Using budding yeast as a model, we study how specialized multi-protein chromatin modifying complexes in this organism perform their physiological functions using biochemical and structural biology approaches.
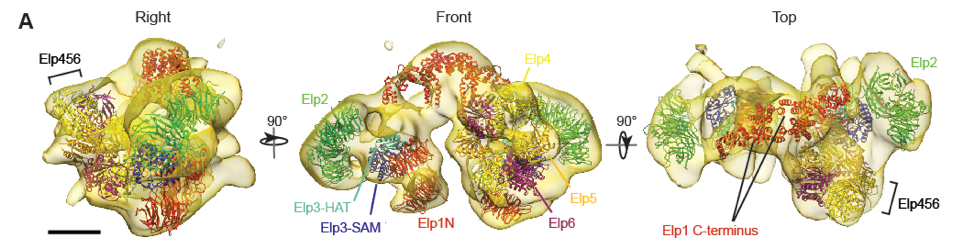
3. Rare genetic diseases
There are over 7,000 different types of rare diseases. It is estimated that 1 in 12 Canadians is affected by a rare disease. Although the genetic bases of many of these diseases have been uncovered, the molecular mechanism of how these genetic changes lead to severe clinical phenotypes remains undefined. We are applying biochemical, structural, and cell biology approaches to address this problem, with an emphasis on the severe multi-system disorder Vici syndrome (https://vicisyndrome.org).
Publications
Comprehensive List
Selected publications:
- Harris NJ, Jenkins ML, Nam SE, Rathinaswamy MK, Parson MAH, Ranga-Prasad H, Dalwadi U, Moeller BE, Sheeky E, Hansen SD, Yip CK, Burke JE. Allosteric activation or inhibition of PI3Kgamma mediated through conformational changes in the p110gamma helical domain. (2023) eLife 12:RP88058.
- Dalwadi U, Corrado E, Fleming KD, Moeller BE, Nam SE, Burke JE, Yip CK. Biochemical characterization of the TINTIN module of the NuA4 complex reveals allosteric regulation of nucleosome interaction. (2022) Molecular & Cellular Biology 42: e0017022.
- Dalwadi U, Mannar D, Zierhut F, Yip CK. Biochemical and structural characterization of human core Elongator and its subassemblies. (2022) ACS Omega 7: 3424-3433.
- Rathinaswamy MK*, Dalwadi U*, Fleming KD, Adams C, Stariha JJTB, Pardon E, Baek M, Vadas O, DiMaio F, Steyaert J, Hansen SD, Yip CK+, Burke JE+. Structure of the phosphoinositide 3-kinase (PI3K) p110gamma-p101 complex reveals molecular mechanism of GPCR activation. (2021) Science Advances 7: eabj4282.
- Nam SE, Cheung YWS, Nguyen TN, Gong M, Chan S, Lazarou M, Yip CK. Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5. (2021) Communications Biology 4: 291.
- Cheung YWS, Nam SE, Yip CK. Recent advances in single-particle electron microscopic analysis of autophagy degradation machinery. (2020) International Journal of Molecular Biosciences. 21: 8051.
- Setiaputra D, Ahmad S, Dalwadi U, Steunou AL, Lu S, Ross JD, Dong MQ, Cote J, Yip CK. Molecular architecture of the essential yeast histone acetyltransferase complex NuA4 redefines its multimodularity. (2018) Molecular & Cellular Biology 38: 300570-17
- Nanji T, Liu X, Chew LH, Li FK, Biswas M, Yu ZQ, Lu S, Dong MQ, Du LL, Klionsky DJ, Yip CK. (2017) Conserved and unique features of the fission yeast core Atg1 complex. (2017) Autophagy 13: 2018-2027.
- Setiaputra D, Cheng DT, Lu S, Hansen JH, Dalwadi U, Lam CH, To JL, Dong MQ, Yip CK. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangment. (2017) EMBO Reports 18: 280-291.
*authors contributed equally; +co-corresponding authors