Rose Zhou – Defended MSc Thesis

Congratulations Rose! On June 4th, 2021 Rose Zhou successfully defended her thesis, “Investigating the in vitro and in vivo anti-resorptive effects of herbal-and TCM-based extracts on Cathepsin K activity.” Rose was a trainee in Dr. Dieter Bromme’s Lab.
Enhancing the understanding of how diseases occur in one organ but not another
An international team led by UBC researchers has used proteomics to map how proteins interact, revealing how the same protein, expressed in two different tissues, can have dramatically different impacts.

“These findings have significantly advanced the understanding of how the same set of protein “parts” can be differently arranged in cells across tissues,” says first author Dr. Nicollas Scott, a former UBC fellow who now heads a laboratory at the Doherty Institute in Melbourne, Australia.
“This is a major advance for basic science, but also in our understanding of human disease,” says senior author Dr. Leonard Foster, a professor in UBC’s Department of Biochemistry and Molecular Biology. “Many inherited diseases are caused by a genetic mutation that is present in every cell in the body, but causes dysfunction in only one tissue.”
In humans and other life forms, proteins are encoded by the genome and interact with one another to perform normal cellular functions. “Proteins are like parts and although there is only a limited number of different types of parts in a given cell, how these can be put together in organizations known as protein complexes, can be quite different,” explains first author Michael Skinnider, a UBC MD/PhD student based in Dr. Foster’s lab at the LSI.
In the two decades since the human genome was first sequenced, vast amounts of money have been poured into mapping complete protein-protein interaction networks in humans and other model organisms.

Up until now, most insights into protein interaction networks have been gained using cell culture-based systems, but these do not always mirror what is observed within tissues.
“As a result, their relevance to physiological contexts like living tissues has never been truly clear,” states senior author Dr. Jörg Gsponer, associate professor in UBC’s Department of Biochemistry and Molecular Biology. For example, inherited cardiac myopathies cause problems in the heart, but not the liver or the thymus. Understanding the differences that exist between different tissues in the human body will help clarify why certain diseases occur in one organ and not another.”
“By characterizing how the protein complexes of tissues are put together, this can help us explain the functionality differences between tissues and why disease associated proteins can have certain impacts in one tissue over another,” adds Scott.
“Using these findings, we have generated a resource for the community so that people can look at what each protein interacts with in different tissues to gain new insights into different disease models and better understand how a given protein works in their authentic states,” said Skinnider.
These protein-protein interaction maps were built using a novel technique called protein correlation profiling, which enabled the team to take protein complexes isolated from mouse tissues, separate them using size exclusion chromatography, then monitor which proteins are found together. Then by using computational approaches and the concept of ‘guilt by association’ the team reconstructed the protein interaction networks in each tissue.

“Being able to use tissues to explore interaction networks enables us to get a better picture of how proteins are interacting with each other in a way which is a lot closer to what is actually happening in our own bodies,” highlights Scott.
“Understanding the molecular organization of all the different cells in the different tissues in the human body has been, and is of central interest to many branches of life science,” reflects Foster. “As has so often been the case, technological developments from our labs enabled us to take on this challenge.” (Kristensen et al., Nature Methods 2012).
Over the past decade, the researchers have refined and adapted both the wet-lab techniques as well as the sophisticated computational tools that are required to make sense of the huge amount of data their technique generates. “However, bringing the assay into in vivo mice was a major technological leap that occurred in this study for the first time, and allowed us to take a major step forward towards understanding proteins and their interactions in physiological contexts,” says Gsponer.
“Our hope is that this will be a high-quality resource to be used by thousands of research groups around the world,” adds Skinnider. “This is the first data where we have been able to directly measure the interaction network in different tissues, as opposed to just predicting what turned out to be low quality networks.”

The research team plans to continue to develop the technology, and are currently applying it to study, among other things, how the interaction network in honey bees responds to infection, and how the interaction network in human cells responds to a coronavirus, such as SARS-CoV-2.
Read the paper:
Skinnider M.A., Scott, N.E., Prudova, A. , Craig H. Kerr, C.H., Stoynov, N., Stacey, R.G. et al. (2021) An atlas of protein-protein interactions across mouse tissues DOI: 10.1016/j.cell.2021.06.003.
This post originally appeared on the LSI Website.
Calem Kenward wins 2021/22 Killam Doctoral Award

Calem Kenward, a PhD student in the lab of Dr. Natalie Strynadka has been named a 2021/22 recipient of a Killam Doctoral Scholarship.
His work on proteins has included exploring the crystal structure of the main protease of SARS-CoV-2.
In an interview with UBC Graduate studies, Calem said he has always loved taking things apart and figuring out how they worked. “I like to think working in a research lab is the ultimate version of that. In science you get to drill down to the tiniest, most fundamental parts of a system and see how they interact to create incredible complexity around us. In some small way, I hope to contribute to our understanding of that complexity.
“I study the pinnacle of microscopic machinery, that is the proteins that compose almost all living things,” he said. “These little machines are composed of the simplest repeating sub-units but work to make machines thousands of times more efficient and smaller than we can artificially. Understanding how these proteins function and interact is a significant challenge, and UBC has the tools (and connections) to make studying these challenging targets possible.”
Working in the Strynadka lab, he has been able to pursue the opportunity to get involved in cutting-edge research and expand his abilities as a biochemist. During the pandemic, the Strynadka lab has been working on the crystalline structure of the SARS-CoV-2 virus, producing multiple papers using cutting edge techniques. “Science is always changing at a rapid pace,” said Calem, “but it feels like that pace is increasing more than ever. Staying on top of all the advancements in techniques and methods while contending with the global competition is going to be a significant challenge.”
This post originally appeared on the LSI Website.
Translating discovery to transform health: Untold stories from medical research
*Webinar Series: Transforming Health for Everyone Webinar Series – Brought to you by UBC Faculty of Medicine Development & Alumni Engagement
The rapid pace of development for COVID-19 treatments and vaccines has demonstrated both the need and opportunity to expedite discoveries from the lab to the clinic—where innovation can save and improve lives.
British Columbia is home to a thriving life sciences ecosystem that made pivotal contributions to the COVID-19 response. The biotech sector in B.C. is poised to drive both the economy and healthcare of the future—yet individually, scientists, companies and clinicians still face seemingly insurmountable barriers to collaboration and innovation. Can you imagine a smoother path?
Hear from UBC experts Dr. Pieter Cullis, Dr. Megan Levings and Dr. Sriram Subramaniam as they share insights from their own experiences bringing discoveries into the healthcare system, and perspectives on how to integrate academic research and the biotech industry in B.C. to accelerate health innovation.
Wednesday, July 14, 2021
11:00 a.m. – 12:30 p.m. PDT
Click here to register
Speakers

Dr. Pieter R. Cullis, BSc’67, MSc‘70, PhD’72 is a Professor in the Department of Biochemistry and Molecular Biology at UBC and Scientific Director and CEO of NanoMedicines Innovation Network, Canada’s National Centre of Excellence in Nanomedicines. Dr. Cullis and collaborators have been responsible for fundamental advances in the design and development of nanomedicines employing lipid nanoparticle technology for cancer therapies and gene therapies. This work has contributed to five drugs that have been approved by regulatory agencies in the US, Europe and Canada. Dr. Cullis has co-founded 10 biotechnology companies that now employ over 300 people, has published over 350 scientific articles and is an inventor on over 60 patents. He also co-founded the Centre for Drug Research and Development, a Centre of Excellence for the Commercialization of Research (now AdMare) in 2004, the Personalized Medicine Initiative in 2012 and the NanoMedicines Innovation Network in 2019. Dr. Cullis was elected a Fellow of the Royal Society of Canada in 2004 and was also awarded the Prix Galien, Canada’s premier prize for achievements in pharmaceutical R&D, in 2011.
Two recently approved drugs, enabled by lipid nanoparticle delivery systems devised by Dr. Cullis, members of his UBC laboratory and colleagues in the companies he has co-founded, deserve special emphasis. The first is Onpattro, which was approved by the US FDA in August 2018 to treat the previously fatal hereditary condition transthyretin-induced amyloidosis. Onpattro is the first RNAi drug to receive regulatory approval. The second is BNT162b2, the COVID-19 vaccine developed by Pfizer/BioNTech that has been approved in Canada, the USA, the UK and Europe. It is anticipated that more than 1.3billion doses of BNT162b2 will be administered worldwide in 2021.

Dr. Megan Levings, PhD’99 is a Professor in the Department of Surgery and School of Biomedical Engineering at UBC and Lead of the Childhood Diseases Theme at the BC Children’s Hospital Research Institute. Her lab studies how a special kind of white blood cell, known as a T regulatory cell, could be used as a cellular therapy to stop harmful immune responses. She leads a vibrant group of trainees and staff who are researching how to use T regulatory cells to replace conventional immunosuppression in the context of transplantation and autoimmunity. She has won numerous awards, including the Canadian Society for Immunology Investigator Award and the YWCA Woman of Distinction, Science, Research and Technology. She is internationally recognized in the field of human immunology and currently chairs the Federation of Clinical Immunology Societies Centers of Excellence.

Dr. Sriram Subramaniam received his PhD in Physical Chemistry from Stanford University and completed postdoctoral training in the Departments of Chemistry and Biology at the Massachusetts Institute of Technology. He is currently a Professor of Biochemistry and Molecular Biology and Urologic Sciences and the Gobind Khorana Chair in Cancer Drug Design at UBC. Before arriving at UBC, Dr. Subramaniam was Senior Investigator and Chief of the Biophysics Section at the US National Cancer Institute, where he founded and directed the Center for Molecular Microscopy and the National Cryo-EM Facility.
Dr. Subramaniam has made seminal contributions in several areas of modern biological electron microscopy. Work in his research group has shown that cryo-EM methods can be used to evaluate drug binding, at atomic resolution, to clinically relevant targets, which has the potential to revolutionize the landscape of drug discovery. Other contributions he has made include structural studies of membrane proteins using electron crystallography, in the development of electron tomographic methods for the study of HIV, influenza and related viruses for vaccine development, and in pioneering new focused ion beam-based technologies for cell and tissue imaging. Dr. Subramaniam leads an interdisciplinary program at UBC that aims to catalyze transformative discoveries in cancer, neuroscience and infectious disease and to create the next generation technologies and therapies that will have a lasting impact on global health.
Moderator

Kathryn Gretsinger, MJ’06 is an Associate Professor of teaching at the UBC School of Journalism, Writing and Media, Senior Faculty Advisor to the UBC President and Director of the Global Reporting Centre at UBC. She is a long time public broadcaster at the Canadian Broadcasting Corporation, with a record of creating award-winning work at the local and national level. Kathryn is also a Killam Teaching Prize winner, and she was named as one of North America’s top innovative journalism educators in 2018. Kathryn’s scholarship focuses on interdisciplinary learning, including an advisory role with the Lind Initiative, a position as a faculty member in residence at UBC’s Emerging Media Lab, and providing instruction in faculties across UBC from the School of Population and Public Health (Drugs and Society) to Science (Communicating Science) to work with scholars from the Peter Wall Institute.
Find the original post on the Faculty of Medicine Website.
Xinying Wang – Defended MSc Thesis
 Congratulations Amy! On May 20th, 2021 Amy Wang successfully defended her thesis, “Exploring the evolution of a viral internal ribosome entry site”. Amy have been a trainee in Dr. Eric Jan’s Lab.
Congratulations Amy! On May 20th, 2021 Amy Wang successfully defended her thesis, “Exploring the evolution of a viral internal ribosome entry site”. Amy have been a trainee in Dr. Eric Jan’s Lab.
Covid-19 vaccines have triggered the next wave of pharmaceuticals – Dr. Pieter Cullis
 Quartz Magazine’s Katherine Ellen Foley has interviewed Dr. Pieter Cullis in a wide-ranging story exploring the dawning age of genetic medicine.
Quartz Magazine’s Katherine Ellen Foley has interviewed Dr. Pieter Cullis in a wide-ranging story exploring the dawning age of genetic medicine.
“Covid-19 vaccines are the start of a new wave of genetic medicine—drugs that tweak DNA to keep us healthy,” begins Foley’s piece, which asserts, “chances are, you’ve never heard of Pieter Cullis. But if you’ve received the Pfzer-BioNTech or Moderna Covid-19 vaccines, you have him, in part, to thank.”
Foley’s article is posted behind a paywall on Quartz Magazine’s website. It can be accessed via a free trial, here: https://qz.com/2005658/covid-19-vaccines-have-ushered-in-the-age-of-genetic-medicine/.
This post originally appeared on the LSI Website.
Congratulations to Rachel Price on winning a Dragon’s Den Award
Rachel Price is a recipient of BCREGMED‘s Dragon’s Den Award.
Rachel is a PhD candidate in the Teves Lab at UBC. In the lab, she studies the action of transcription factors during mitosis in mouse embryonic stem cells to investigate the mechanisms of stem cell self-renewal and differentiation. Outside of the lab, she enjoys knitting, hiking, and baking sourdough.
Read about the BCREGMED’s Dragon’s Den Award: event details

UBC researchers unveil first molecular images of B.1.1.7 COVID-19 mutation
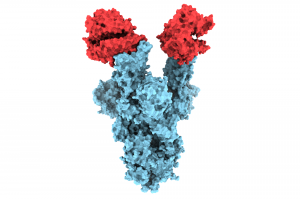
Using cryo-electron microscopy, UBC researchers have revealed the structure of the N501Y spike protein mutant, shown above (in blue) bound to two copies of the ACE2 receptor (in red).
UBC researchers are the first in the world to publish structural images of the N501Y mutation on the SARS-CoV-2 spike protein—a change believed to be partly responsible for the highly infectious nature and rapid spread of variant B.1.1.7.
The pictures, taken at near-atomic resolution, provide critical insight as to why the B.1.1.7 variant—first detected in the U.K and now accounting for a growing number of cases across Canada—is more infectious. The images also add to the growing body of data indicating that existing vaccines are likely to remain effective in preventing mild and severe cases caused by B.1.1.7.
Dr. Sriram Subramaniam, professor in UBC faculty of medicine’s department of biochemistry and molecular biology, discusses the implications of his team’s research, recently published in PLOS Biology.
What do these images reveal about the B.1.1.7 variant?
Viruses are constantly mutating. The B.1.1.7 variant of concern, which was first reported to the World Health Organization in mid-December 2020, has an unusually large number of mutations. Of particular interest is a mutation known as N501Y located on the virus’s spike protein, which is what the virus uses to attach itself to human cells.
The images we captured provide the first structural glimpse of the N501Y mutant and show that the changes resulting from the mutation are localized. In fact, the N501Y mutation is the only mutation in the B.1.1.7 variant that is located on the portion of the spike protein that binds to the human ACE2 receptor, which is the enzyme on the surface of our cells that serves as the entry gate for SARS-CoV-2.
Will existing vaccines remain effective?
Our analysis revealed that even though the N501Y mutant can bind and enter our cells more readily, it can still be neutralized by antibodies that block the entry of the unmutated version of the virus into cells.
This is an important observation and adds to the growing body of evidence that the majority of antibodies elicited in our immune system by existing vaccines are likely to remain effective in protecting us against the B1.1.7 variant.
How did you capture these structural images?
The SARS-CoV-2 virus is 100,000 times smaller than the size of a pinhead, making it undetectable using a regular light microscope. The proteins on the surface of a virus are even smaller.
To visualize the detailed shapes of viruses and proteins, we use cryo-electron microscopes, which can be up to 12 feet tall. This powerful imaging technology uses beams of electrons to visualize shapes of tissues and cells using ultra-cooling, or “cryo” techniques—essentially, the imaging of samples at liquid nitrogen temperatures.
Are you researching other COVID variants?
We are currently examining other variants, including P.1 (Brazilian), B.1.351 (South African), B.1.427/B.1.429(Californian) and B.1.617 (Indian) variants, and trying to understand how these mutations alter how the spike protein interacts with neutralizing antibodies. We are also looking at how these mutations may change how the virus binds to ACE2.
It’s important to understand the different molecular structures of these emerging variants to determine whether they’ll respond to existing treatments and vaccines and ultimately find ways to control their spread more effectively.
Find the Original Post: UBC News
Rachel Jun – Defended MSc Thesis
 Congratulations Rachel! On April 19, 2021 Hee Jin Rachel Jun successfully defended her thesis, “Exploring the role of mutations in the signal peptides of VIM-2, NDM-1, and IMP-1 in the development of advantageous phenotypes in Escherichia coli.” Rachel have been training in Dr. Nobuhiko Tokuriki’s Lab.
Congratulations Rachel! On April 19, 2021 Hee Jin Rachel Jun successfully defended her thesis, “Exploring the role of mutations in the signal peptides of VIM-2, NDM-1, and IMP-1 in the development of advantageous phenotypes in Escherichia coli.” Rachel have been training in Dr. Nobuhiko Tokuriki’s Lab.
